Difference between revisions of "Time Course"
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <big>In the last 6 months, 3 new members of the omicron (B.1.1.529) lineage have emerged, and subsequently been recognized as variants of interest (VOI) by the World Health Organization (WHO), which are the BA.2.75, XBB, and BQ.1 subvariants that rose to prominence | + | <big>In the last 6 months, 3 new members of the omicron (B.1.1.529) lineage have emerged, and subsequently been recognized as variants of interest (VOI) by the World Health Organization (WHO), which are the BA.2.75, XBB, and BQ.1 subvariants that rose to prominence between July and October 2022. Each of these VOIs has brought along an array of novel mutable sites crucial for refining the viral fitness of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Leading mutations identified by our deLemus analysis that emerged within the aforementioned timeframe are listed as follows:<br /></big> |
| + | |||
| + | <tabs> | ||
| + | <tab name="2022.08.31"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/32.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | </tab> | ||
| + | |||
| + | <tab name="2022.06.30"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/30.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | </tab> | ||
| + | </tabs> | ||
| + | |||
| + | <!-- | ||
==2022.08.31== | ==2022.08.31== | ||
<htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/32.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/32.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| Line 28: | Line 41: | ||
| N460K || BA.2.75 | | N460K || BA.2.75 | ||
|} | |} | ||
| + | --> | ||
| + | |||
==2021.03.31== | ==2021.03.31== | ||
<htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/15.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/15.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
Revision as of 10:55, 14 February 2023
In the last 6 months, 3 new members of the omicron (B.1.1.529) lineage have emerged, and subsequently been recognized as variants of interest (VOI) by the World Health Organization (WHO), which are the BA.2.75, XBB, and BQ.1 subvariants that rose to prominence between July and October 2022. Each of these VOIs has brought along an array of novel mutable sites crucial for refining the viral fitness of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Leading mutations identified by our deLemus analysis that emerged within the aforementioned timeframe are listed as follows:
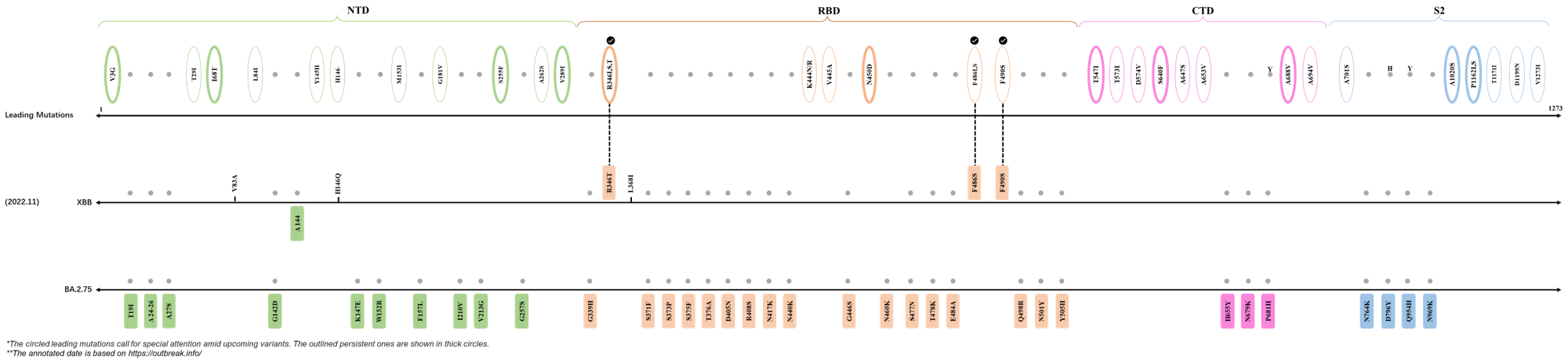

2021.03.31
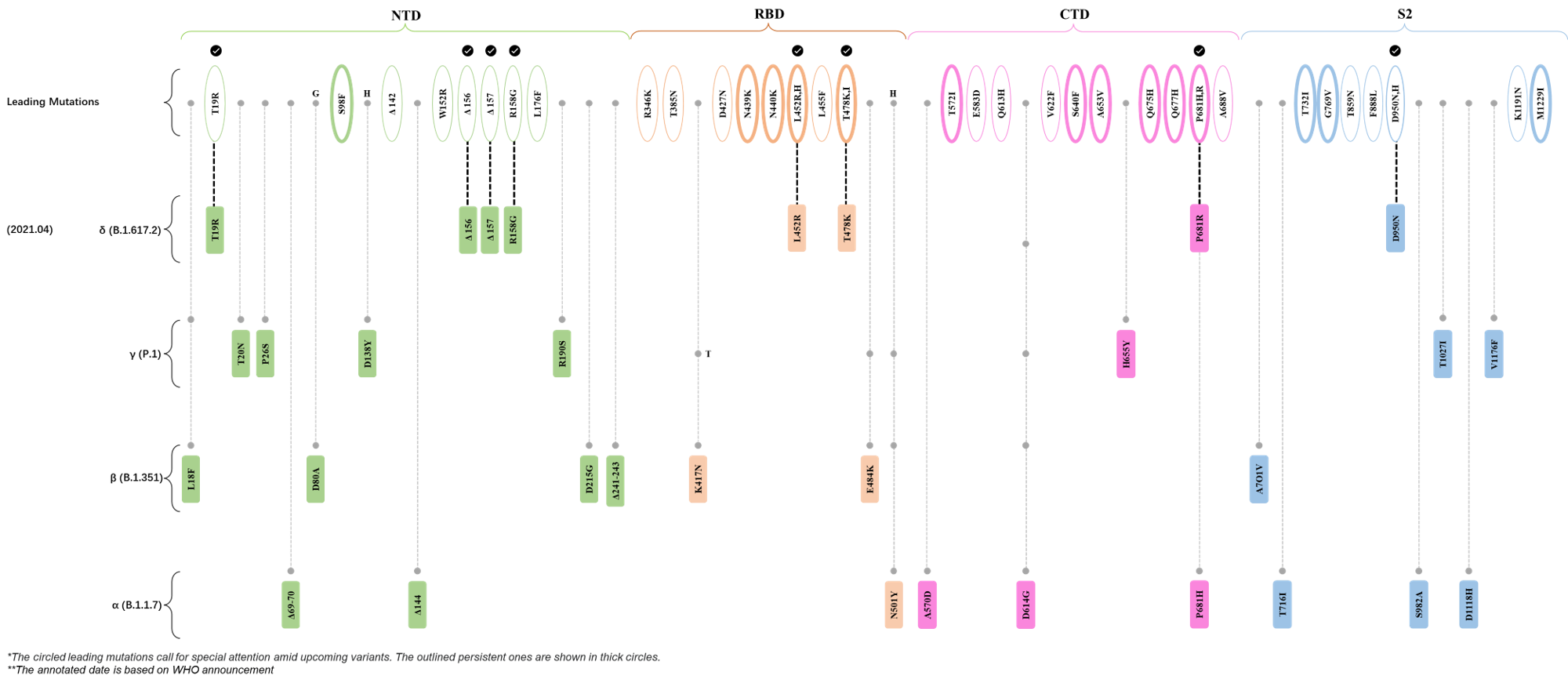
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| T19R | δ (B.1.617.2) |
| E156- | δ (B.1.617.2) |
| F157- | δ (B.1.617.2) |
| R158G | δ (B.1.617.2) |
| L452R | δ (B.1.617.2) |
| T478K | δ (B.1.617.2) |
| P681R | δ (B.1.617.2) |
| D950N | δ (B.1.617.2) |
2020.10.31
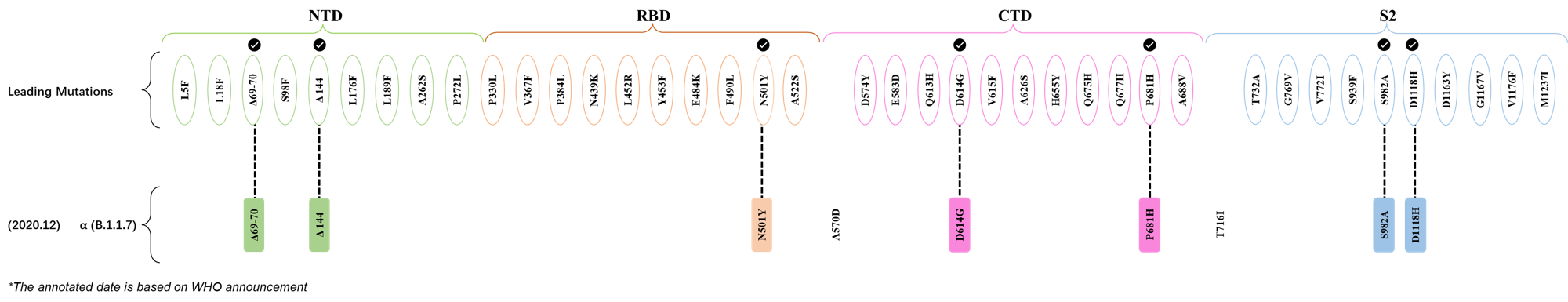
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H69- | α (B.1.1.7) |
| V70- | α (B.1.1.7) |
| Y144- | α (B.1.1.7) |
| N501Y | α (B.1.1.7) |
| D614G | α (B.1.1.7) |
| P681H | α (B.1.1.7) |
| S982A | α (B.1.1.7) |
| D1118H | α (B.1.1.7) |