Difference between revisions of "deLemus"
| Line 2: | Line 2: | ||
''Dynamic Expedition of Leading Mutations in SARS-CoV-2 Spike Glycoproteins'' | ''Dynamic Expedition of Leading Mutations in SARS-CoV-2 Spike Glycoproteins'' | ||
</br> | </br> | ||
| − | + | <!-- | |
The dynamic epidemiology of coronavirus disease 2019 (COVID-19) since its outbreak has been a result of the continuous evolution of its etiological agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Within the first 2 years of this pandemic, the World Health Organization (WHO) has already announced 4 variants of concern (VOC), namely alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2), together with numerous variants of interest (VOI). The latest lineage to be designated a VOC would be omicron (B.1.1.529),<ref name="Karim" /> from which a diverse variant soup is generated.<ref>Callaway, E. COVID ‘variant soup’ is making winter surges hard to predict. ''Nature'' '''611,''' 213 (2022).</ref> From the original BA.1 strain of November 2021 to the most recent XBB and BQ.1 strains of late 2022,<ref name="Wang" /><ref name="European Centre" /> each omicron subvariant has successively proliferated and outcompeted its once dominant antecedent.<ref name="Del Rio" /> The emergence of all these variants has brought along many novel mutations that continue to fine-tune the fitness of the virus,<ref>Carabelli, A. M. ''et al.'' SARS-CoV-2 variant biology: Immune escape, transmission and fitness. ''Nat Rev Microbiol'' (2023). DOI: https://doi.org/10.1038/s41579-022-00841-7.</ref><ref>Witte, L. ''et al.'' Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. ''Nat Commun'' '''14,''' 302 (2023).</ref> leading to its persistent global circulation. Recent emerging variant (EV) data retrieved from GISAID, as of 17 January 2023, has revealed that the top 4 most rapidly spreading lineages are the BA.1.1.22, CH.1.1, XBB.1.5, and BQ.1.1 variants, among which XBB.1.5 has been found to be especially prevalent in the US,<ref>Callaway, E. Coronavirus variant XBB.1.5 rises in the United States — is it a global threat? ''Nature'' '''613,''' 222 (2023).</ref> making up of more than 40% of its sequence coverage in early January 2023. | The dynamic epidemiology of coronavirus disease 2019 (COVID-19) since its outbreak has been a result of the continuous evolution of its etiological agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Within the first 2 years of this pandemic, the World Health Organization (WHO) has already announced 4 variants of concern (VOC), namely alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2), together with numerous variants of interest (VOI). The latest lineage to be designated a VOC would be omicron (B.1.1.529),<ref name="Karim" /> from which a diverse variant soup is generated.<ref>Callaway, E. COVID ‘variant soup’ is making winter surges hard to predict. ''Nature'' '''611,''' 213 (2022).</ref> From the original BA.1 strain of November 2021 to the most recent XBB and BQ.1 strains of late 2022,<ref name="Wang" /><ref name="European Centre" /> each omicron subvariant has successively proliferated and outcompeted its once dominant antecedent.<ref name="Del Rio" /> The emergence of all these variants has brought along many novel mutations that continue to fine-tune the fitness of the virus,<ref>Carabelli, A. M. ''et al.'' SARS-CoV-2 variant biology: Immune escape, transmission and fitness. ''Nat Rev Microbiol'' (2023). DOI: https://doi.org/10.1038/s41579-022-00841-7.</ref><ref>Witte, L. ''et al.'' Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. ''Nat Commun'' '''14,''' 302 (2023).</ref> leading to its persistent global circulation. Recent emerging variant (EV) data retrieved from GISAID, as of 17 January 2023, has revealed that the top 4 most rapidly spreading lineages are the BA.1.1.22, CH.1.1, XBB.1.5, and BQ.1.1 variants, among which XBB.1.5 has been found to be especially prevalent in the US,<ref>Callaway, E. Coronavirus variant XBB.1.5 rises in the United States — is it a global threat? ''Nature'' '''613,''' 222 (2023).</ref> making up of more than 40% of its sequence coverage in early January 2023. | ||
| − | + | --> | |
==Spike Glycoprotein== | ==Spike Glycoprotein== | ||
Revision as of 15:37, 23 August 2023
Dynamic Expedition of Leading Mutations in SARS-CoV-2 Spike Glycoproteins
Spike Glycoprotein
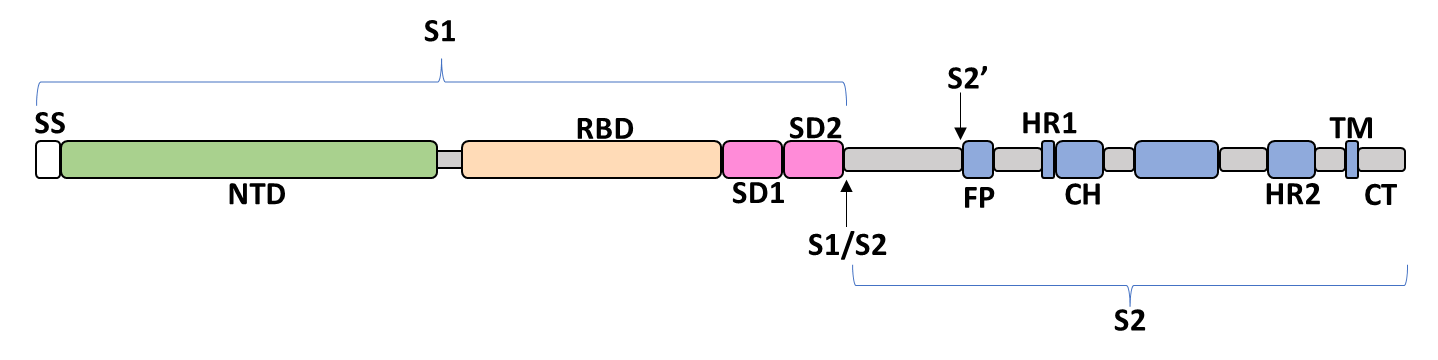
The spike glycoprotein of SARS-CoV-2 is a trimeric type I viral fusion protein that binds the virus to the angiotensin-converting enzyme 2 (ACE2) receptor of a host cell.[1] It is composed of 2 subunits: the N-terminal subunit 1 (S1) and C-terminal subunit 2 (S2), within which multiple domains lie. The S1 region facilitates ACE2 binding and is made up of an N-terminal domain (NTD), a receptor-binding domain (RBD), and 2 C-terminal subdomains (CTD1 and CTD2), while the downstream S2 region is responsible for mediating virus-host cell membrane fusion.
Update
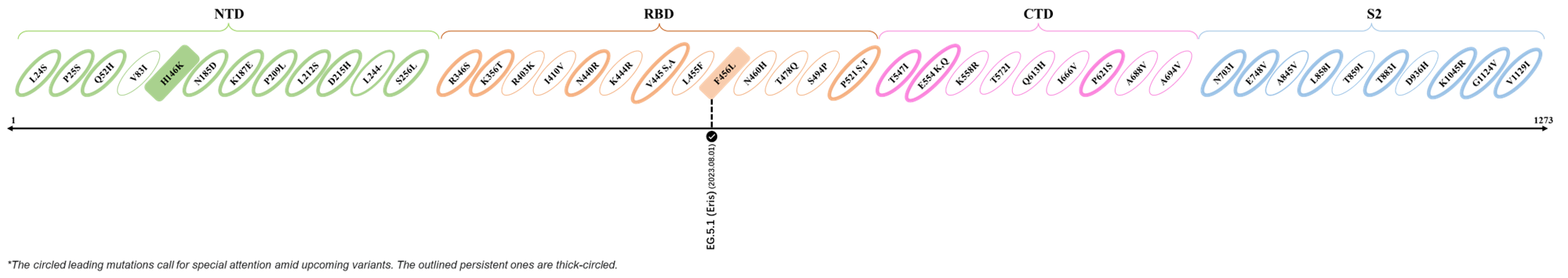
The identified leading mutations in 2023 are listed as follows [2]:
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.08.04 - 2023.08.22
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| N185D | XBB.1.5 |
| L212S | FY.4.2 |
| V445A | XBC.1.6 |
| L455F | EG.5.1.1 |
| F456L | EG.5.1 (Eris) |
| E554Q | XBB.1.5.18 |
| Q613H | XBB.1.16 |
| T883I | XBB.1.16 |
*The reported mutations of detected variants are from Cov-Lineages[3]
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.06.30 - 2023.07.05
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H146K | FL.2.3 (XBB.1.9.1.2.3) |
| S446N | FL.19 |
| F456L | XBF |

- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
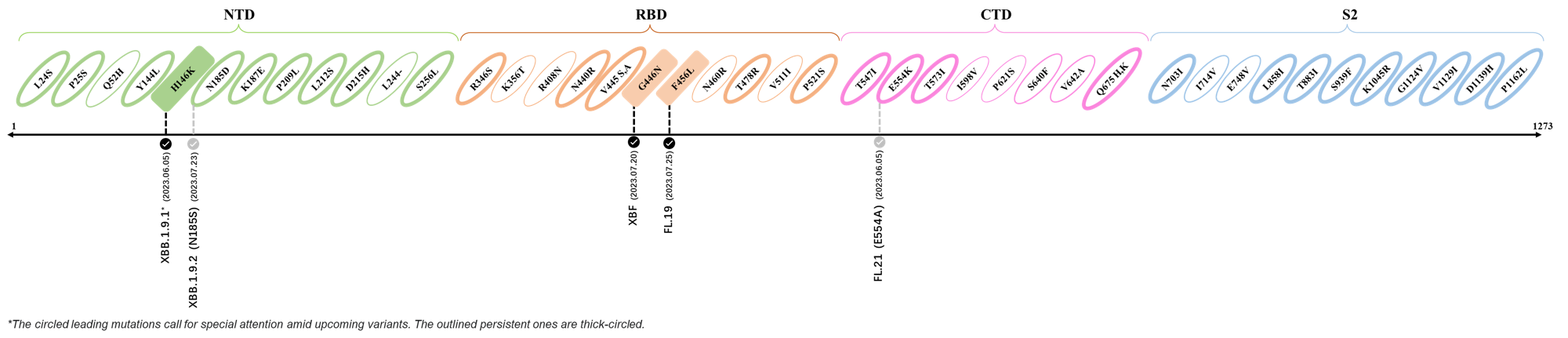
2023.06.01 - 2023.06.13
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| F490P | XBB.1.9.1 |
| E554K | XBB.1.9.1 (sublineage) |
| Q675K | XBB.1.22.1 |
| L858I | CH.1.1.1 |

- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.05.01 - 2023.05.12
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| F456L | FD.1.1 & EG.5.1 (2023.08) |
| S494P | XBB.2.3 & XBB.1.1 |
| T572I | FY.1 ( XBB.1.22.1.1 ) |
*The reported mutations of detected variants are from GISAID

- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.04.01 - 2023.04.21
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H146K | XBB.1.5 & XBB.1.16 |
| M153I | XBB.2.3.3 |
| E180V | XBB.1.16 |
| K444R | XBB.1.5 |
| T478R | XBB.1.16, XBB.1.5, CH.1.1.2 & XBB.2.3 |
| F490P | XBB.2.6 |
| S494P | XBB.1.5 |
| Q613H | XBB.1.16 |
| P621S | XBB.2.3 |
| A688V | XAY.1.1.1 |
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
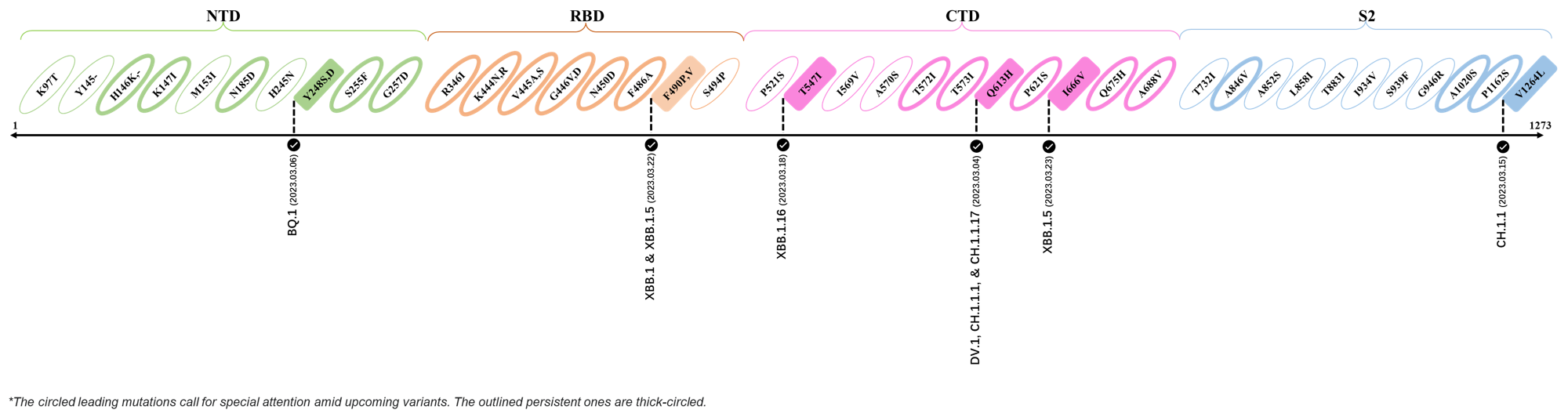
2023.03.01 - 2023.03.21
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| Y248S | BQ.1 |
| F490P | XBB.1 & XBB.1.5 |
| T547I | XBB.1.16 |
| Q613H | DV.1, CH.1.1.1 & CH.1.1.17 |
| I666V | XBB.1.5 |
| V1264L | CH.1.1 |
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
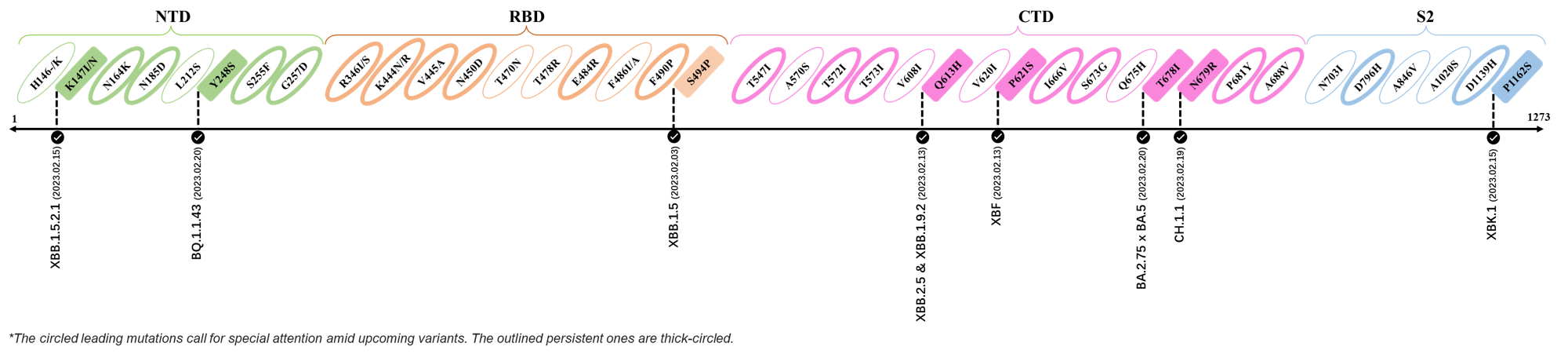
2023.02.03 - 2023.02.20
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| K147I | XBB.1.5.2.1 |
| Y248S | BQ.1.1.43 |
| S494P | XBB.1.5 |
| Q613H | XBB.1.9.2 & XBB.2.4 |
| P612S | XBF |
| T678I | BA.2.75 x BA.5 |
| N679R | CH.1.1 |
| P1162S | XBK.1 |
*The reported mutations of detected variants are from GISAID[4]
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
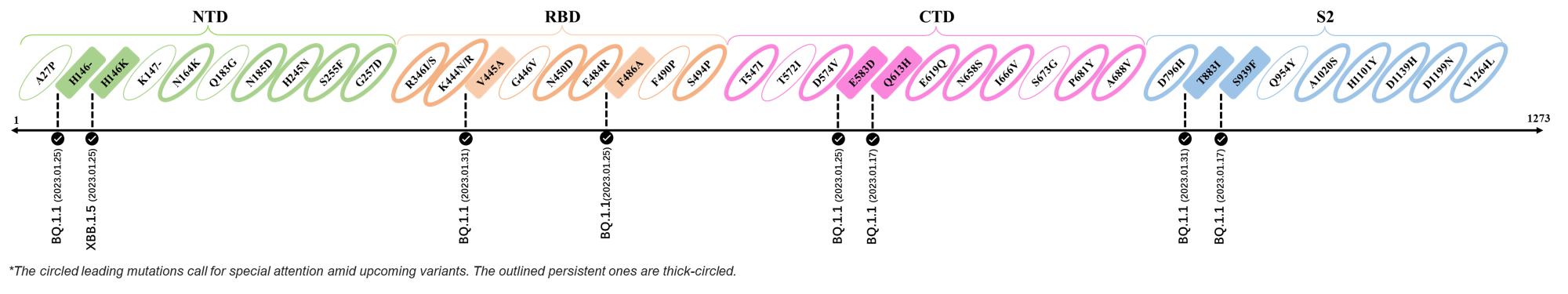
2023.01.31
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| V445A | BQ.1.1 |
| T883I | BQ.1.1 |
2023.01.17 - 2023.01.25
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H146- / H146K | BQ.1.1 / XBB.1.5 |
| F486A | BQ.1.1 |
| E583D | BQ.1.1 |
| Q613H | BQ.1.1 |
| S939F | BQ.1.1 |
References
- ↑ Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23, 3 (2021).
- ↑ deLemus team, Analysis of Leading Mutations in SARS-CoV-2 Spike Glycoproteins (in preparation, 2023).
- ↑ Cov-Lineages https://cov-lineages.org/
- ↑ GISAID https://gisaid.org/
Cite error: <ref> tag with name "Del Rio" defined in <references> is not used in prior text.
Cite error: <ref> tag with name "European Centre" defined in <references> is not used in prior text.
Cite error: <ref> tag with name "Karim" defined in <references> is not used in prior text.
Cite error: <ref> tag with name "Wang" defined in <references> is not used in prior text.
