Difference between revisions of "deLemus"
| (789 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | __NOTOC__ | |
| + | ''Dynamic Expedition of Leading Mutations in SARS-CoV-2 Spike Glycoproteins'' | ||
| − | The | + | </br> |
| + | The dynamic epidemiology of coronavirus disease 2019 (COVID-19) since its outbreak has been a result of the continuous evolution of its etiological agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Within the first 2 years of this pandemic, the World Health Organization (WHO) has already announced 4 variants of concern (VOC), namely alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2), together with numerous variants of interest (VOI). The latest lineage to be designated a VOC would be omicron (B.1.1.529),<ref name="Karim" /> from which a diverse variant soup is generated.<ref>Callaway, E. COVID ‘variant soup’ is making winter surges hard to predict. ''Nature'' '''611,''' 213 (2022).</ref> From the original BA.1 strain of November 2021 to the most recent XBB and BQ.1 strains of late 2022,<ref name="Wang" /><ref name="European Centre" /> each omicron subvariant has successively proliferated and outcompeted its once dominant antecedent.<ref name="Del Rio" /> The emergence of all these variants has brought along many novel mutations that continue to fine-tune the fitness of the virus,<ref>Carabelli, A. M. ''et al.'' SARS-CoV-2 variant biology: Immune escape, transmission and fitness. ''Nat Rev Microbiol'' (2023). DOI: https://doi.org/10.1038/s41579-022-00841-7.</ref><ref>Witte, L. ''et al.'' Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. ''Nat Commun'' '''14,''' 302 (2023).</ref> leading to its persistent global circulation. Recent emerging variant (EV) data retrieved from GISAID, as of 17 January 2023, has revealed that the top 4 most rapidly spreading lineages are the BA.1.1.22, CH.1.1, XBB.1.5, and BQ.1.1 variants, among which XBB.1.5 has been found to be especially prevalent in the US,<ref>Callaway, E. Coronavirus variant XBB.1.5 rises in the United States — is it a global threat? ''Nature'' '''613,''' 222 (2023).</ref> making up of more than 40% of its sequence coverage in early January 2023. | ||
| + | <!-- | ||
| + | --> | ||
| + | ==Spike Glycoprotein== | ||
| + | The spike glycoprotein of SARS-CoV-2 is a trimeric type I viral fusion protein that binds the virus to the angiotensin-converting enzyme 2 (ACE2) receptor of a host cell.<ref name="Jackson2021"/> It is composed of 2 subunits: the N-terminal subunit 1 (S1) and C-terminal subunit 2 (S2), within which multiple domains lie. The S1 region facilitates ACE2 binding and is made up of an N-terminal domain (NTD), a receptor-binding domain (RBD), and 2 C-terminal subdomains (CTD1 and CTD2), while the downstream S2 region is responsible for mediating virus-host cell membrane fusion. | ||
| − | < | + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/Domains.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag> |
| + | =='''Update'''== | ||
| + | The identified leading mutations in 2023 are listed as follows <ref name="deLemus" />: | ||
| − | + | <tabs> | |
| − | {| class="wikitable | + | <tab name="2023.12"> |
| − | + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-12.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | |
| − | + | ||
| − | + | ===2023.12.01-2023.12.17=== | |
| − | + | {| class="wikitable" | |
| − | |||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''L455F'''</span> || EG.5.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''A475V'''</span> || EG.5.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''E654K'''</span> || HK.3 |
| − | | | + | |} |
| − | + | ||
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.11"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-11.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | ===2023.11.01-2023.11.17=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''N185D'''</span> || HK.3.2 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''L455F'''</span> || EG.5.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''A475V'''</span> || JF.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''T572I'''</span> || FY.2 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || XBB.1.16 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''D1153Y'''</span> || HK.3 |
| − | | | + | |} |
| − | + | ||
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.10"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-10.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | ===2023.10.06=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''L455F'''</span> || EG.5.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''A475V'''</span> || GK.1 |
| − | | | + | |} |
| − | + | ||
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.09"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-09.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | ===2023.09.08-2023.09.28=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''R403K'''</span> || BA.2.86 (Pirola) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''L455F'''</span> || EG.5.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''S494P'''</span> || EG.5.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''P521S'''</span> || XBB.1.16.15 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''E554K'''</span> || BA.2.86 (Pirola) & FE.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || BA.2.86 (Pirola) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''P621S'''</span> || BA.2.86 (Pirola) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''T732I'''</span> || XBB.2.3 x XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''S939F'''</span> || BA.2.86 (Pirola) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''V1264L'''</span> || CK.1.1 |
| − | | | + | |} |
| − | + | ||
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.08"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-08.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.08.04 - 2023.08.22=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''N185D'''</span> || XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''L212S'''</span> || FY.4.2 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''V445A'''</span> || XBC.1.6 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''L455F'''</span> || EG.5.1.1 |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F456L'''</span> || EG.5.1 (Eris) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''E554Q'''</span> || XBB.1.5.18 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || XBB.1.16 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''T883I'''</span> || XBB.1.16 |
| − | |The | + | |} |
| − | + | ''*The reported mutations of detected variants are from Cov-Lineages<ref name="Cov-Lineages" />'' | |
| − | | | + | </br> |
| + | ===<big>RBD Mutation Profile of Latest VOIs.</big>=== | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-08_VarRBD.png" alt="test for htmltag img" class="wikimg" style="display: block;width:65%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | </tab> | ||
| + | |||
| + | <tab name="2023.07"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-07.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-7" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-7', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_2023_07.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.06.30 - 2023.07.05=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''H146K'''</span> || FL.2.3 (XBB.1.9.1.2.3) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''S446N'''</span> || FL.19 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F456L'''</span> || XBF |
| − | | | + | |} |
| − | + | ||
| − | | | + | |
| + | </tab> | ||
| + | |||
| + | <tab name="2023.06"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-06.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-6" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-6', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_2023_06.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.06.01 - 2023.06.13=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F490P'''</span> || XBB.1.9.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''E554K'''</span> || XBB.1.9.1 (sublineage) |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q675K'''</span> || XBB.1.22.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''L858I'''</span> || CH.1.1.1 |
| − | | | + | |} |
| − | + | ||
| − | | | + | |
| + | </tab> | ||
| + | |||
| + | <tab name="2023.05"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-05.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-5" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-5', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_2023_05.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.05.01 - 2023.05.12=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F456L'''</span> || FD.1.1 & EG.5.1 (2023.08) |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''S494P'''</span> || XBB.2.3 & XBB.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''T572I'''</span> || FY.1 ( XBB.1.22.1.1 ) |
| − | | | + | |} |
| − | + | ''*The reported mutations of detected variants are from GISAID'' | |
| − | | | + | |
| + | |||
| + | </tab> | ||
| + | |||
| + | <tab name="2023.04"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-04.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-4" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-4', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_2023_04.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.04.01 - 2023.04.21=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''H146K'''</span> || XBB.1.5 & XBB.1.16 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''M153I'''</span> || XBB.2.3.3 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''E180V'''</span> || XBB.1.16 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''K444R'''</span> || XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''T478R'''</span> || XBB.1.16, XBB.1.5, CH.1.1.2 & XBB.2.3 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F490P'''</span> || XBB.2.6 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''S494P'''</span> || XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || XBB.1.16 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''P621S'''</span> || XBB.2.3 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''A688V'''</span> || XAY.1.1.1 |
| − | | | + | |} |
| − | + | ||
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.03"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-03.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-3" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-3', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_2023_03.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.03.01 - 2023.03.21=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''Y248S'''</span> || BQ.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F490P'''</span> || XBB.1 & XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''T547I'''</span> || XBB.1.16 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || DV.1, CH.1.1.1 & CH.1.1.17 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''I666V'''</span> || XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''V1264L'''</span> || CH.1.1 |
| − | | | + | |} |
| − | + | ||
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.02"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-02.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-2" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-2', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_2023_02.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.02.03 - 2023.02.20=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''K147I'''</span> || XBB.1.5.2.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''Y248S'''</span> || BQ.1.1.43 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''S494P'''</span> || XBB.1.5 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || XBB.1.9.2 & XBB.2.4 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''P612S'''</span> || XBF |
| − | |||
| − | |||
| − | | | ||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''T678I'''</span> || BA.2.75 x BA.5 |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''N679R'''</span> || CH.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''P1162S'''</span> || XBK.1 |
| − | | | + | |} |
| − | + | ''*The reported mutations of detected variants are from GISAID<ref name="GISAID" />'' | |
| − | | | + | </tab> |
| + | |||
| + | <tab name="2023.01"> | ||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/2023-01.png" alt="test for htmltag img" class="wikimg" style="display: block;width:100%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | <html> | ||
| + | <style> | ||
| + | .molstar { | ||
| + | position: relative; | ||
| + | width: 80%; | ||
| + | padding-bottom: 56.25%; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="https://molstar.org/viewer/molstar.css" /> | ||
| + | <script type="text/javascript" src="https://molstar.org/viewer/molstar.js"></script> | ||
| + | |||
| + | <div id="viewer-1" class="molstar" style="display: block; margin-left:auto; margin-right:auto; padding-bottom: 40%;"></div> | ||
| + | <script type="text/javascript"> | ||
| + | molstar.Viewer.create('viewer-1', { | ||
| + | layoutIsExpanded: false, | ||
| + | layoutShowControls: false, | ||
| + | layoutShowRemoteState: false, | ||
| + | layoutShowSequence: true, | ||
| + | layoutShowLog: false, | ||
| + | layoutShowLeftPanel: true, | ||
| + | |||
| + | viewportShowExpand: true, | ||
| + | viewportShowSelectionMode: false, | ||
| + | viewportShowAnimation: false, | ||
| + | }).then(viewer => { | ||
| + | viewer.loadSnapshotFromUrl('https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/pdb/LM_latest.molx', 'molx'); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | * Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022). | ||
| + | |||
| + | <big>Here are the recently confirmed leading mutations.</big> | ||
| + | |||
| + | ===2023.01.31=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''V445A'''</span> || BQ.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''T883I'''</span> || BQ.1.1 |
| − | | | + | |} |
| − | + | ===2023.01.17 - 2023.01.25=== | |
| − | | | + | {| class="wikitable" |
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:yellowgreen;">'''H146- / H146K'''</span> || BQ.1.1 / XBB.1.5 |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:burlywood;">'''F486A'''</span> || BQ.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''E583D'''</span> || BQ.1.1 |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:hotpink;">'''Q613H'''</span> || BQ.1.1 |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | <span style="color:cornflowerblue;">'''S939F'''</span> || BQ.1.1 |
| − | | | + | |} |
| − | | | + | |
| − | | | + | </tab> |
| + | |||
| + | </tabs> | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | ===2023.01.31=== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants !! Conformation | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | V445A || BQ.1.1 || Amino acid site located at an RBD epitope<ref name="Weisblum_eLife"/> ; Mutation reduces neutralization by antibody <ref name="CellRep20220517"/> |
| − | | | + | |} |
| − | | | + | ===2023.01.17 - 2023.01.25=== |
| − | | | + | {| class="wikitable" |
|- | |- | ||
| − | + | ! Outlined Mutations !! Confirmed in VOC/Emerging Variants !! Conformation | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | H146-/K || BQ.1.1, XBB.1.5 || Amino acid site recognized by mAbs targeting NTD<ref name=":3"/> |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | E583D || BQ.1.1 || Viral functions to be confirmed by further investigation |
| − | | | ||
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | | Q613H || BQ.1.1 || Speculate to enhance replicative fitness and transmissibility due to close proximity to D614G ; Potential functions to be elucidated<ref name=":0"/><ref name="Bugembe"/> |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | S939F || BQ.1.1 || Destabilize both pre-fusion and post-fusion S2 conformation<ref name="Olivie"/> ; Capable to enhance infectivity and modulate T-cell immune response when combined with D614G<ref name="LiImpactCell"/><ref name="Donzelli"/> |
| − | | | + | |} |
| − | | | + | |
| − | | | + | <big>The following leading mutations call for special attention with respect to the upcoming variants.</big> |
| + | ==NTD== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Conformation | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | A27P || An antigenic site targeted by the group 3 antibody C1717<ref name=":2" /> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | K147- || Involved in interacting with multiple monoclonal antibodies<ref name=":4" /> ; Mutation to threonine (K147T) at this site promotes immune evasion<ref name=":3" /> |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | N164K || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | Q183G || Interactions with surface glycoconjugates mediate the viral attachment<ref name="Sun_Glycobio2021" /> ; Caused a loss of an amide group; May abrogate the hydrogen bond between the amino acid and the carboxylic group of surface sialosides<ref name="Buchanan" /> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | N185D || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | |H245N |
| − | | | + | |Located in the supersite loop of the NTD antigenic supersite for antibodies SLS28 and S2X333<ref name=":4" /><ref name=":3" /> ; Caused a loss of a positive charge ; Introduces an NXS sequon (<sub>245</sub>NRS<sub>247</sub>) for ''N''-glycosylation |
| − | |||
| − | |||
|- | |- | ||
| − | | | + | |G252V |
| − | | | + | |Site is critical for the binding of human antibody COV2-3439<ref>Suryadevara N. ''et al.'' An antibody targeting the N-terminal domain of SARS-CoV-2 disrupts the spike trimer. ''J Clin Invest'' '''132,''' 159062 (2022).</ref> |
| − | |||
| − | |||
|- | |- | ||
| − | | | + | |G257D |
| − | | | + | |Located in the supersite loop of the NTD antigenic supersite for antibodies SLS28 and S2X333<ref name=":4" /><ref name=":3" /> ; Caused a gain of negative charge |
| − | |||
| − | |||
|- | |- | ||
| − | | | + | |A262S |
| − | | | + | |Enhance the utilization of ACE2 in numerous mammals<ref name="Wang_JMedVirol2022" /> ; May increase interspecies and intraspecies transmissibility |
| − | | | + | |} |
| − | | | + | |
| + | ==RBD== | ||
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Conformation | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | R346I/S || Possibly lead to immune evasion due to the disruption of class 3 antibodies binding site<ref name="Gaebler"/> <ref name="WangQ_LancetID2022"/> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | K444N/R || Escape mutations for covalescent plasma<ref name="Weisblum_eLife"/> |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | G446V || Substantially decreases the neutralization titers of plasma<ref name="Greaney"/> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | N450D || Results in antibody resistance<ref name="Cong_CellHM2021"/> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | E484R/S || A site of mutation being reported in multiple variants, mutation at this site could harbor escape mutations that impede the binding and neutralization ability of antibodies<ref name=":0"/> <ref name="Greaney"/> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | F490P || Mutation at this site enables antibody escape over mAb COV2-2479, COV2-2050, COV2-2096 based on DMS study.<ref name="Greaney"/> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | S494P || This mutation persistently shows up in an immunocompromised patient of COVID-19, which was treated various drugs and antibodies e.g. remdesivir, intravenous immunoglobulin, etc.<ref name="Choi"/> |
| − | | | + | |} |
| − | + | ||
| − | | | + | ==CTDs== |
| + | {| class="wikitable" | ||
|- | |- | ||
| − | + | ! Outlined Mutations !! Conformation | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | T547I || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | T572I || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | D574V || Located at the CTD1 region, substitution to an electrically neutral valine residue permits the endosomal entry efficiency and immune evasion ability of SARS-CoV-2.<ref name="Zhou_CellHM2020"/> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | E619Q || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | E658S || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | I666V || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | S673G || Functional impact to be confirmed in future investigation. |
| − | | | ||
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | P681Y || Located at the C-terminal of the CTD2, this substitution can diminish the cleavage efficiency of the S1/S2 interface because the bulky nature of tyrosine hinders the binding of furin to the cleavage loop.<ref name="Henrich"/><ref name="Tian_2009"/></big> |
| − | | | ||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | I688V || Functional impact to be confirmed in future investigation. |
| − | | | + | |} |
| − | | | + | ==S2== |
| − | | | + | {| class="wikitable" |
|- | |- | ||
| − | + | ! Outlined Mutations !! Conformation | |
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | D796H || Located in S2 region, the single aspartic acid-to-histidine substitution was found to enhance the neutralization resistance of the spike glycoprotein in a chronical infection patient.<ref name="KempCIP" /></big> |
| − | | | + | |} |
| − | + | ||
| − | | | + | == References == |
| + | <references> | ||
| + | <ref name="XBB.1.5">Yue, C. ''et al''. Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. Preprint at https://www.biorxiv.org/content/10.1101/2023.01.03.522427v2 (2023).</ref> | ||
| + | <ref name=":4">Cao, Y. ''et al.'' Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. ''Nature'' (2022). DOI:10.1038/s41586-022-05644-7</ref> | ||
| + | <ref name="Zahradník">Zahradník, J. ''et al.'' SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. ''Nat Microbiol'' '''6,''' 1188 (2021).</ref> | ||
| + | <ref name="Makowski">Makowski, E. K., Schardt, J. S., Smith, M. D. & Tessier, P. M. Mutational analysis of SARS-CoV-2 variants of concern reveals key tradeoffs between receptor affinity and antibody escape. ''PLOS Comput Biol'' '''18,''' (2022).</ref> | ||
| + | <ref name=":0">Qu, P. ''et al.'' Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. ''Cell Host Microbe'' '''30,''' 1518 (2022).</ref> | ||
| + | <ref name=":2">Tamura, T. ''et al.'' Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two omicron subvariants. Preprint at https://www.biorxiv.org/content/10.1101/2022.12.27.521986v1 (2022).</ref> | ||
| + | <ref name=":3">Wang, Q. ''et al.'' Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. ''Cell'' '''186,''' 279 (2023).</ref> | ||
| + | <ref name=":1">Qu, P. ''et al.'' Enhanced neutralization resistance of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. ''Cell Host Microbe'' '''31,''' 9 (2023).</ref> | ||
| + | <ref name="Tuekprakhon">Tuekprakhon, A. ''et al.'' Antibody escape of SARS-CoV-2 omicron BA.4 and BA.5 from Vaccine and BA.1 Serum. ''Cell'' '''185,''' 2422 (2022).</ref> | ||
| + | <ref name="Wang">Wang, Q. ''et al.'' Antibody evasion by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4 and BA.5. ''Nature'' '''608,''' 603 (2022).</ref> | ||
| + | </references> | ||
| + | |||
| + | ==Summary== | ||
| + | <tabs> | ||
| + | <tab name="NTD"><htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/NTD.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag></tab> | ||
| + | <tab name="RBD"><htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/RBD.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag></tab> | ||
| + | <tab name="CTDs"><htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/CTDs.png" alt="test for htmltag img" class="wikimg" style="display:block;width:70%;margin-left: auto;margin-right: auto;"></htmltag></tab> | ||
| + | <tab name="S2"><htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/S2.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag></tab> | ||
| + | </tabs> | ||
| + | |||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/NTD.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/RBD.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/CTDs.png" alt="test for htmltag img" class="wikimg" style="display:block;width:70%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | <htmltag tagname="img" src="https://wiki.laviebay.hkust.edu.hk/deLemus/RESEARCH_TEAMS/images/PublishedPlot/S2.png" alt="test for htmltag img" class="wikimg" style="display: block;width:70%;margin-left: auto;margin-right: auto;"></htmltag> | ||
| + | |||
| + | == '''Deep Mutational Scanning Data''' == | ||
| + | <big>The RBD-ACE2 binding data</big><ref>Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE Jr, Bloom JD. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021 Jan 13;29(1):44-57.e9. doi: 10.1016/j.chom.2020.11.007. Epub 2020 Nov 19. PMID: 33259788; PMCID: PMC7676316.</ref> <big>showed that R346S, N354S, E484R and S494P are the mutations lead to increased binding affinity in all the 5 background sequence.</big> | ||
| + | {| class="wikitable" | ||
| + | |+ | ||
| + | RBD-ACE2 binding affinity | ||
| + | |'''Unique Mutations''' | ||
| + | |'''Date''' | ||
| + | |'''Wuhan''' | ||
| + | |'''Alpha''' | ||
| + | |'''Beta''' | ||
| + | |'''Eta''' | ||
| + | |'''Delta''' | ||
|- | |- | ||
| − | | | + | |'''R346S''' |
| − | | | + | |2023.01 |
| − | | | + | |0.12 |
| − | | | + | |0.14 |
| + | |0.07 | ||
| + | |0.03 | ||
| + | |0.11 | ||
|- | |- | ||
| − | | | + | |'''N354S''' |
| − | | | + | |2023.05 |
| − | | | + | |0.03 |
| − | | | + | |0.01 |
| + | |0.04 | ||
| + | |0.32 | ||
| + | |0.02 | ||
|- | |- | ||
| − | | | + | |'''E484R''' |
| − | | | + | |2023.01 |
| − | | | + | |0.06 |
| − | | | + | |0.04 |
| + | | - | ||
| + | | - | ||
| + | |0.11 | ||
|- | |- | ||
| − | | | + | |'''S494P''' |
| − | | | + | |2023.01 |
| − | | | + | |0.33 |
| − | | | + | |0.18 |
| − | | | + | |0.13 |
| − | | | + | |0.14 |
| − | + | |0.06 | |
| − | + | |} | |
| − | + | <big>Immune escape data</big><ref>Tyler N. Starr., et al., Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution.''Science''377,420-424(2022).DOI:10.1126/science.abo7896</ref> <big>shows that the escape ability of R346S, V445A, G446I, and E484R against certain antibodies exceeds 90% mutations.</big> | |
| − | + | {| class="wikitable" | |
| − | + | |+ | |
| − | + | Immune Escaping | |
| − | + | |'''Unique Mutations''' | |
| − | | | + | |'''Date''' |
| − | | | + | |'''Antybody1''' |
| − | + | |'''Antybody2''' | |
| − | | | + | |'''Antybody3''' |
| − | | | + | |'''Antybody4''' |
| − | + | |'''Antybody5''' | |
| − | | | ||
| − | | | ||
| − | | | ||
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | |'''R346S''' |
| − | | | + | |2023.01 |
| − | + | |COV2-2082 | |
| − | + | |COV2-2096 | |
| − | + | |COV2-2479 | |
| − | + | |COV2-2832 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | |- | ||
| − | | | ||
| − | |||
| − | | | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | | | ||
|- | |- | ||
| − | | | + | |'''V445A''' |
| − | | | + | |2023.01 |
| − | | | + | |COV2-2050 |
| − | | | + | |COV2-2094 |
| + | |COV2-2479 | ||
| + | |COV2-2499 | ||
| + | |COV2-2677 | ||
|- | |- | ||
| − | | | + | |'''G446I''' |
| − | | | + | |2023.05 |
| − | | | + | |COV2-2096 |
| + | |COV2-2479 | ||
| + | |COV2-2499 | ||
| | | | ||
| − | |||
| − | |||
| − | |||
| − | |||
| | | | ||
|- | |- | ||
| − | | | + | |'''E484R''' |
| − | | | + | |2023.01 |
| − | + | |COV2-2050 | |
| − | | | + | |COV2-2096 |
| − | + | |COV2-2479 | |
| − | + | |COV2-2832 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | |||
| − | |- | ||
| − | | | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | | | ||
|} | |} | ||
| + | <big>Overall, by the first half of this year, '''R346S''' and '''E484R''' are the most potential dangerous mutations we captured.</big> | ||
| + | --> | ||
| + | |||
| + | ==References== | ||
| + | <references> | ||
| + | <ref name="Del Rio">Rössler, A. ''et al''. BA.2 and BA.5 Omicron Differ Immunologically from Both BA.1 Omicron and Pre-Omicron Variants. ''Nat Commun'' '''13''', 7701 (2022)</ref> | ||
| + | <ref name="European Centre">Qu, P. ''et al''. Enhanced Neutralization Resistance of SARS-CoV-2 Omicron Subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. ''Cell Host Microbe'' '''31''', 9 (2023)</ref> | ||
| + | <ref name="Jackson2021">Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. ''Nat Rev Mol Cell Biol'' '''23,''' 3 (2021).</ref> | ||
| + | <ref name="Karim">Karim, S. S. A. & Karim, Q. A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. ''Lancet'' '''398,''' 2126 (2021).</ref> | ||
| + | <ref name="Wang">Wang, Q. ''et al.'' Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. ''Cell'' '''186,''' 279 (2023).</ref> | ||
| + | <ref name="deLemus">deLemus team, Analysis of Leading Mutations in SARS-CoV-2 Spike Glycoproteins (in preparation, 2023).</ref> | ||
| + | <ref name="GISAID">GISAID https://gisaid.org/</ref> | ||
| + | <ref name="Cov-Lineages">Cov-Lineages https://cov-lineages.org/</ref> | ||
| + | </references> | ||
| + | |||
| + | |||
| + | <html><a href="https://www.revolvermaps.com/livestats/locations/57gzazn1dbb/"><img src="//rf.revolvermaps.com/h/m/a/0/ff0000/128/0/57gzazn1dbb.png" width="180" height="120" alt="Map" style="border:0;"></a></html> | ||
| + | <html> | ||
| + | <div id="sfc9pt5d2hy2d328lsrbbs6nlkm4sax62ug"></div> | ||
| + | <script type="text/javascript" src="https://counter10.optistats.ovh/private/counter.js?c=9pt5d2hy2d328lsrbbs6nlkm4sax62ug&down=async" async></script> | ||
| + | <noscript><a href="https://www.freecounterstat.com" title="free website counter"><img src="https://counter10.optistats.ovh/private/freecounterstat.php?c=9pt5d2hy2d328lsrbbs6nlkm4sax62ug" border="0" title="free website counter" alt="free website counter"></a></noscript> | ||
| + | </html> | ||
| + | [[Category:deLemus]] | ||
Latest revision as of 10:25, 15 December 2023
Dynamic Expedition of Leading Mutations in SARS-CoV-2 Spike Glycoproteins
The dynamic epidemiology of coronavirus disease 2019 (COVID-19) since its outbreak has been a result of the continuous evolution of its etiological agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Within the first 2 years of this pandemic, the World Health Organization (WHO) has already announced 4 variants of concern (VOC), namely alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2), together with numerous variants of interest (VOI). The latest lineage to be designated a VOC would be omicron (B.1.1.529),[1] from which a diverse variant soup is generated.[2] From the original BA.1 strain of November 2021 to the most recent XBB and BQ.1 strains of late 2022,[3][4] each omicron subvariant has successively proliferated and outcompeted its once dominant antecedent.[5] The emergence of all these variants has brought along many novel mutations that continue to fine-tune the fitness of the virus,[6][7] leading to its persistent global circulation. Recent emerging variant (EV) data retrieved from GISAID, as of 17 January 2023, has revealed that the top 4 most rapidly spreading lineages are the BA.1.1.22, CH.1.1, XBB.1.5, and BQ.1.1 variants, among which XBB.1.5 has been found to be especially prevalent in the US,[8] making up of more than 40% of its sequence coverage in early January 2023.
Spike Glycoprotein
The spike glycoprotein of SARS-CoV-2 is a trimeric type I viral fusion protein that binds the virus to the angiotensin-converting enzyme 2 (ACE2) receptor of a host cell.[9] It is composed of 2 subunits: the N-terminal subunit 1 (S1) and C-terminal subunit 2 (S2), within which multiple domains lie. The S1 region facilitates ACE2 binding and is made up of an N-terminal domain (NTD), a receptor-binding domain (RBD), and 2 C-terminal subdomains (CTD1 and CTD2), while the downstream S2 region is responsible for mediating virus-host cell membrane fusion.
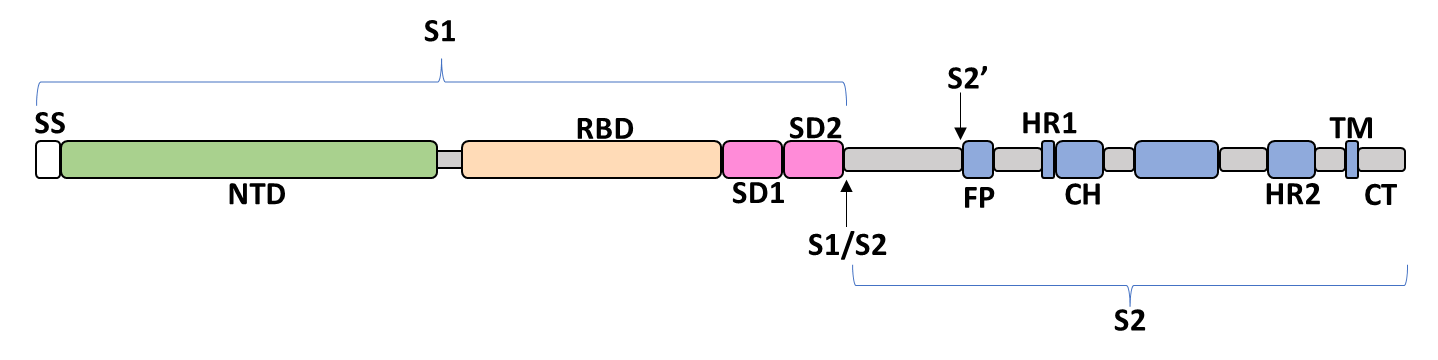
Update
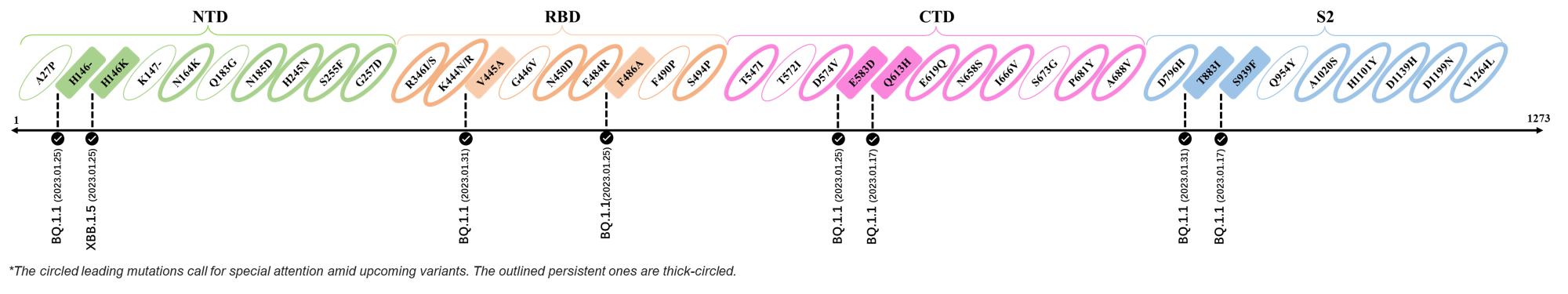
The identified leading mutations in 2023 are listed as follows [10]:
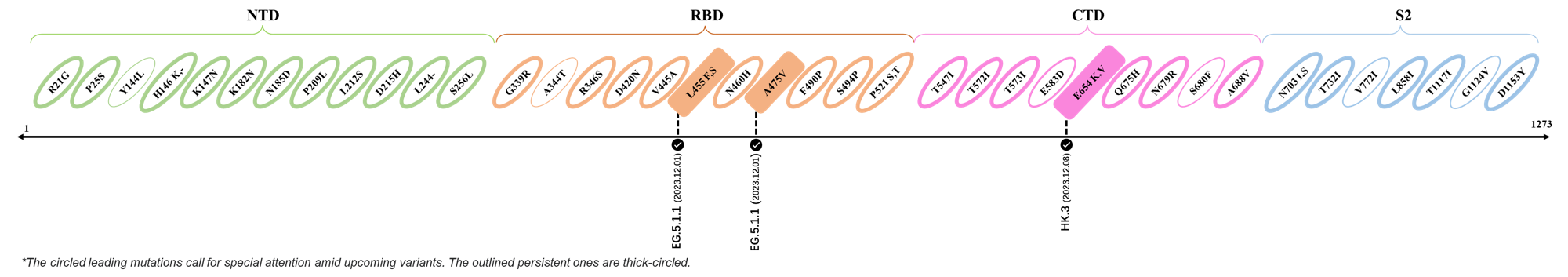
2023.12.01-2023.12.17
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| L455F | EG.5.1.1 |
| A475V | EG.5.1.1 |
| E654K | HK.3 |

2023.11.01-2023.11.17
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| N185D | HK.3.2 |
| L455F | EG.5.1.1 |
| A475V | JF.1 |
| T572I | FY.2 |
| Q613H | XBB.1.16 |
| D1153Y | HK.3 |

2023.10.06
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| L455F | EG.5.1.1 |
| A475V | GK.1 |
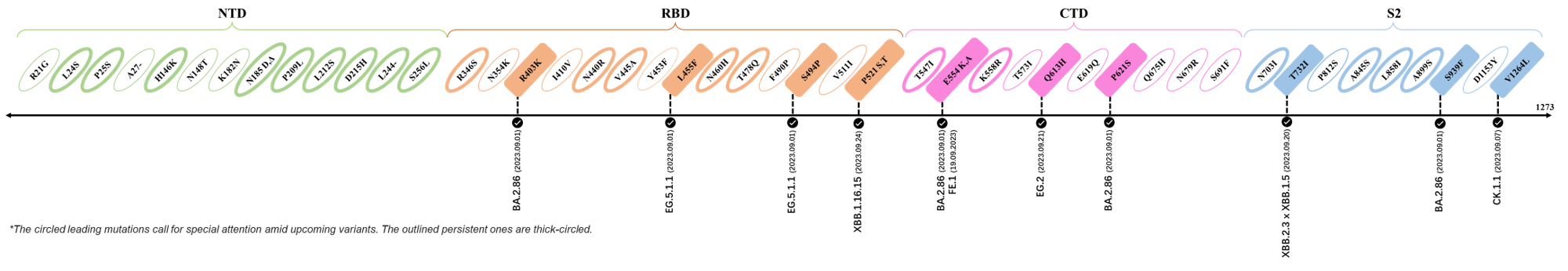
2023.09.08-2023.09.28
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| R403K | BA.2.86 (Pirola) |
| L455F | EG.5.1.1 |
| S494P | EG.5.1.1 |
| P521S | XBB.1.16.15 |
| E554K | BA.2.86 (Pirola) & FE.1 |
| Q613H | BA.2.86 (Pirola) |
| P621S | BA.2.86 (Pirola) |
| T732I | XBB.2.3 x XBB.1.5 |
| S939F | BA.2.86 (Pirola) |
| V1264L | CK.1.1 |
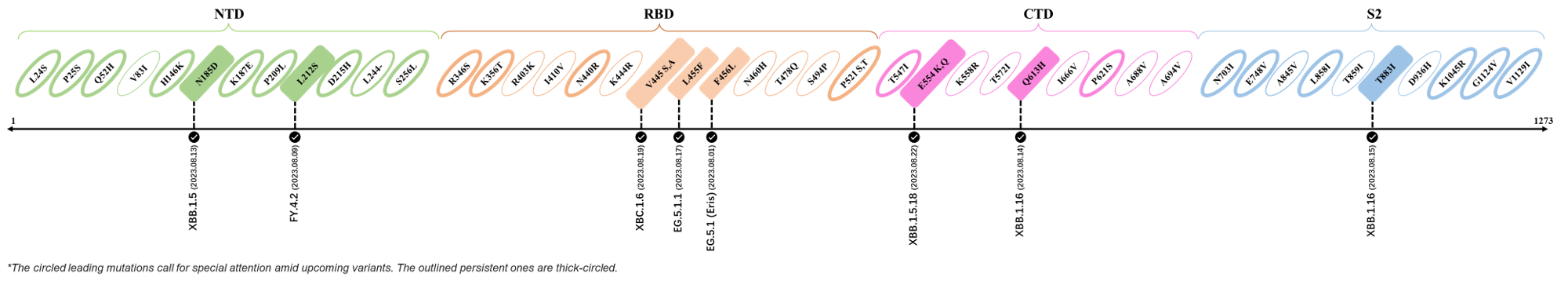
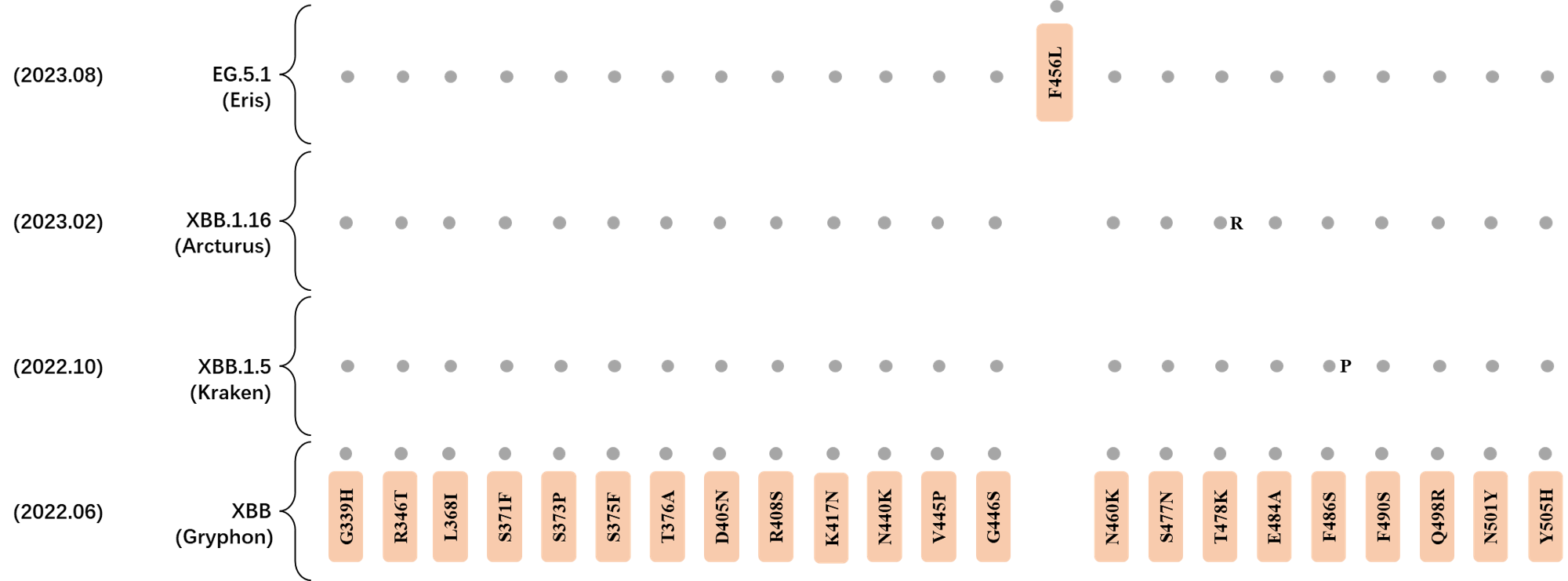
Here are the recently confirmed leading mutations.
2023.08.04 - 2023.08.22
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| N185D | XBB.1.5 |
| L212S | FY.4.2 |
| V445A | XBC.1.6 |
| L455F | EG.5.1.1 |
| F456L | EG.5.1 (Eris) |
| E554Q | XBB.1.5.18 |
| Q613H | XBB.1.16 |
| T883I | XBB.1.16 |
*The reported mutations of detected variants are from Cov-Lineages[11]
RBD Mutation Profile of Latest VOIs.
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.06.30 - 2023.07.05
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H146K | FL.2.3 (XBB.1.9.1.2.3) |
| S446N | FL.19 |
| F456L | XBF |

- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.06.01 - 2023.06.13
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| F490P | XBB.1.9.1 |
| E554K | XBB.1.9.1 (sublineage) |
| Q675K | XBB.1.22.1 |
| L858I | CH.1.1.1 |

- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.05.01 - 2023.05.12
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| F456L | FD.1.1 & EG.5.1 (2023.08) |
| S494P | XBB.2.3 & XBB.1.1 |
| T572I | FY.1 ( XBB.1.22.1.1 ) |
*The reported mutations of detected variants are from GISAID

- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
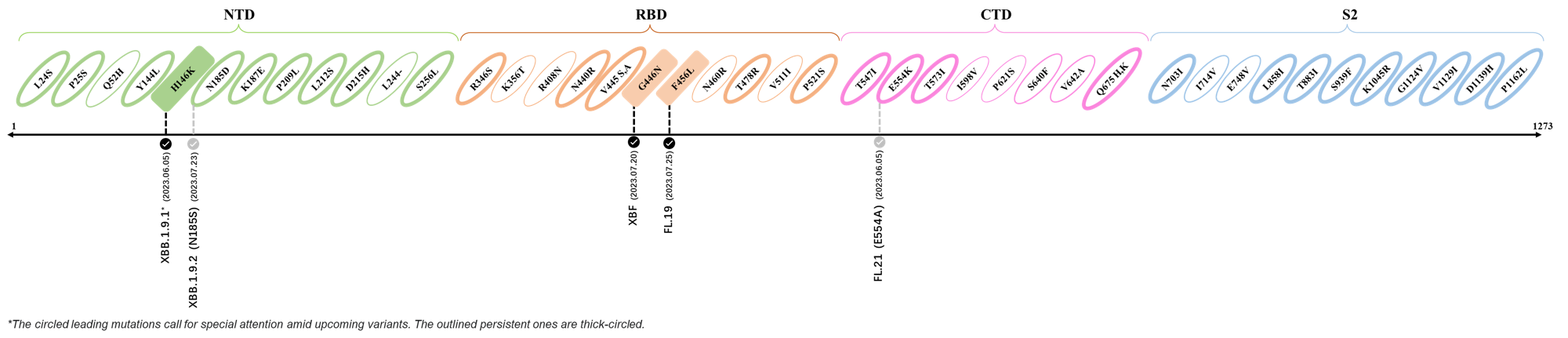
2023.04.01 - 2023.04.21
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H146K | XBB.1.5 & XBB.1.16 |
| M153I | XBB.2.3.3 |
| E180V | XBB.1.16 |
| K444R | XBB.1.5 |
| T478R | XBB.1.16, XBB.1.5, CH.1.1.2 & XBB.2.3 |
| F490P | XBB.2.6 |
| S494P | XBB.1.5 |
| Q613H | XBB.1.16 |
| P621S | XBB.2.3 |
| A688V | XAY.1.1.1 |
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
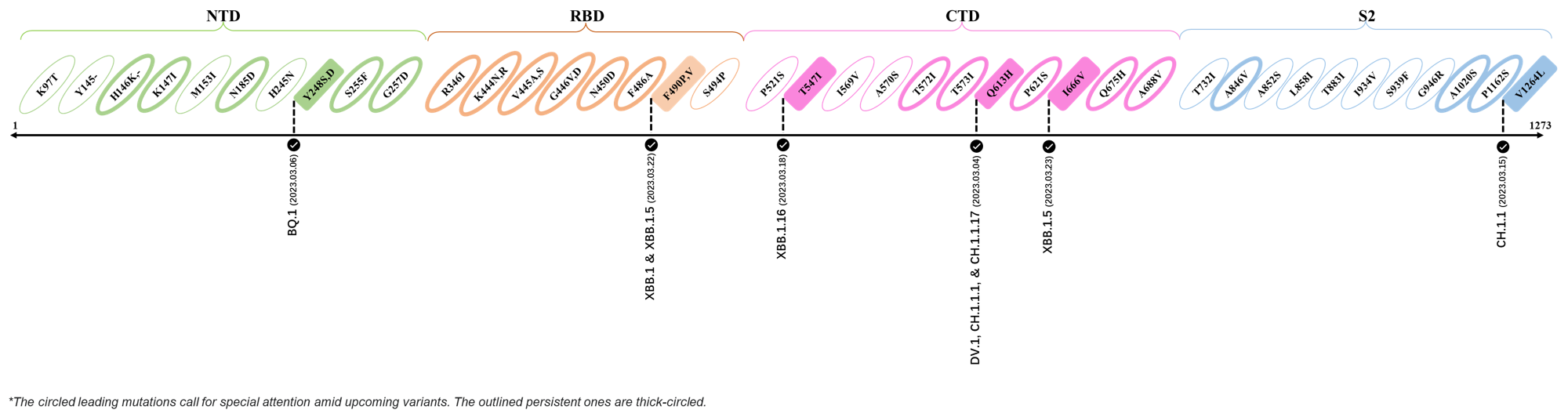
2023.03.01 - 2023.03.21
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| Y248S | BQ.1 |
| F490P | XBB.1 & XBB.1.5 |
| T547I | XBB.1.16 |
| Q613H | DV.1, CH.1.1.1 & CH.1.1.17 |
| I666V | XBB.1.5 |
| V1264L | CH.1.1 |
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
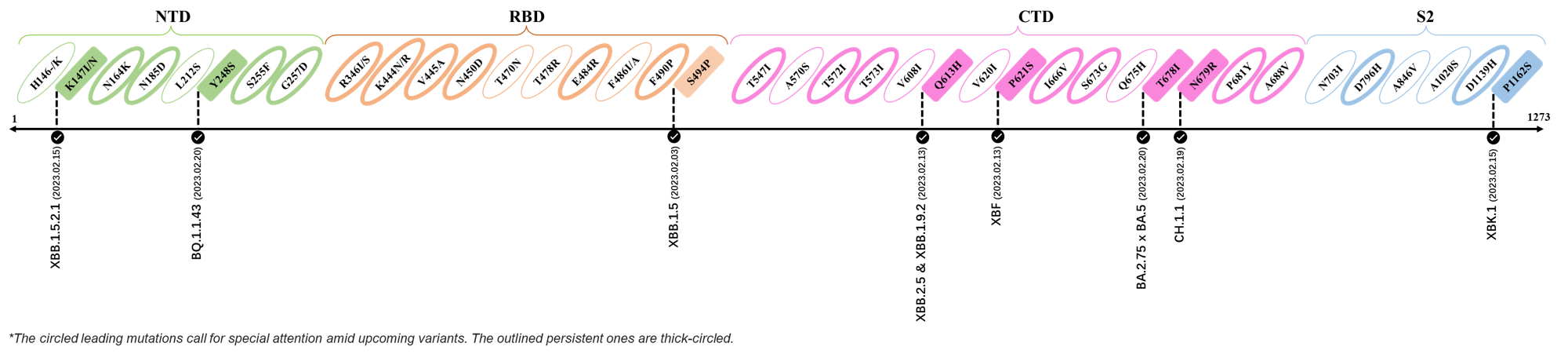
2023.02.03 - 2023.02.20
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| K147I | XBB.1.5.2.1 |
| Y248S | BQ.1.1.43 |
| S494P | XBB.1.5 |
| Q613H | XBB.1.9.2 & XBB.2.4 |
| P612S | XBF |
| T678I | BA.2.75 x BA.5 |
| N679R | CH.1.1 |
| P1162S | XBK.1 |
*The reported mutations of detected variants are from GISAID[12]
- Generated 3D structure of spike protein with highlighted leading mutations (AlphaFold2, colab version 2022).
Here are the recently confirmed leading mutations.
2023.01.31
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| V445A | BQ.1.1 |
| T883I | BQ.1.1 |
2023.01.17 - 2023.01.25
| Outlined Mutations | Confirmed in VOC/Emerging Variants |
|---|---|
| H146- / H146K | BQ.1.1 / XBB.1.5 |
| F486A | BQ.1.1 |
| E583D | BQ.1.1 |
| Q613H | BQ.1.1 |
| S939F | BQ.1.1 |
References
- ↑ Karim, S. S. A. & Karim, Q. A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 398, 2126 (2021).
- ↑ Callaway, E. COVID ‘variant soup’ is making winter surges hard to predict. Nature 611, 213 (2022).
- ↑ Wang, Q. et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279 (2023).
- ↑ Qu, P. et al. Enhanced Neutralization Resistance of SARS-CoV-2 Omicron Subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 31, 9 (2023)
- ↑ Rössler, A. et al. BA.2 and BA.5 Omicron Differ Immunologically from Both BA.1 Omicron and Pre-Omicron Variants. Nat Commun 13, 7701 (2022)
- ↑ Carabelli, A. M. et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat Rev Microbiol (2023). DOI: https://doi.org/10.1038/s41579-022-00841-7.
- ↑ Witte, L. et al. Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. Nat Commun 14, 302 (2023).
- ↑ Callaway, E. Coronavirus variant XBB.1.5 rises in the United States — is it a global threat? Nature 613, 222 (2023).
- ↑ Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23, 3 (2021).
- ↑ deLemus team, Analysis of Leading Mutations in SARS-CoV-2 Spike Glycoproteins (in preparation, 2023).
- ↑ Cov-Lineages https://cov-lineages.org/
- ↑ GISAID https://gisaid.org/
